October 19, 2010
The pandemic Influenza keeps reassorting
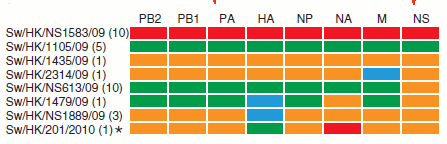
Genome composition of Influenza virus sampled on Hong Kong. Source: Science 2010, reference at the end of the text.
![]() After more than a year of the Influenza A H1N1 episode, the virus is still being monitored all over the world, both the flu cases and genetics diversity of the virus. Following up the genetic diversity helps to understand if the vaccine is still efficient and helps identifying the possible appearance of new strains.
After more than a year of the Influenza A H1N1 episode, the virus is still being monitored all over the world, both the flu cases and genetics diversity of the virus. Following up the genetic diversity helps to understand if the vaccine is still efficient and helps identifying the possible appearance of new strains.
In Hong Kong, location with an important role in the appearance of new viruses because of the bird market and high concentration of people, the virus infected raised pigs. And the sample analysis from June 2009 to February 2010 showed what was already expected: the virus is still reassorting itself.
The reassortement happens when two or more different viruses infect the same cell. The Influenza has eight genes that must be in the viruses that go out so they can be infective, and when more than one virus is present in this cell, the viruses formed are a mosaic of genes from different sources. This process was better explained here. The reassortement is worrisome because, the virus it originates has more abrupt changes than the ones acquired by the most common mutation processes, and these changes can help the virus avoid our immunological system.
In all hosts the reassortement is common. In birds, it was responsible for the creation of the H5N1 virus. In humans, it originated the pandemic viruses H2N2 on 1957 and H3N2 on 1968, and created a third strain of swine Influenza on 1997. Until 1996, there were two main strains of the swine virus: the one called classic that appeared in North American pigs around 1918 and is still there today; the Eurasian virus, also originated
on birds, that circulates in pigs since 1979. On 1997, the two swine viruses reassorted themselves as a new bird virus, originating the strain known as a triple-reassorted virus.
What the Chinese research group found, is well summarized in the figure that illustrates this post. On the left are the name of the viruses found. As it was explained here, the name contains many information. The Sw refers to the host, swine, HK is the location, Hong Kong, the number in bars is the code attributed to the sample, for tabulation, and /09 or 2010 refers to the year it was collected.
Still in the year 2009, three pure virus strains were found in pigs and contained genes of an established strain (they are not pure in fact because many were originated from reassortment events), the human Influenza A H1N1 with its eight genes in red, the Eurasian virus, with its eight genes in green, and the triple-reassorted virus, with eight genes in yellow. There were also found viruses with new reassortement standards both among Eurasian and triple-reassorted kinds, and north American classic, highlighted in blue.
In the beginning of this year, the first reassorted virus with 2009 Influenza A H1N1genes was found. It should not be an isolated problem, given the rate this event occurs and the swine origin of the virus, which already selected competent genes so that the virus multiplies in pigs.
This finding reinforces the need to monitor farm animals, specially birds and swine, to prevent the appearance of new strains and to promote the preparation when needed. Reassortment is a common event, and we are creating more and more animals and traveling around the world easily. While we do not have a vaccine that protects us against various strains of Influenza, prevention still is our best shot.
Source:
Vijaykrishna, D., Poon, L., Zhu, H., Ma, S., Li, O., Cheung, C., Smith, G., Peiris, J., & Guan, Y. (2010). Reassortment of Pandemic H1N1/2009 Influenza A Virus in Swine Science, 328 (5985), 1529-1529 DOI: 10.1126/science.1189132
No Comments » Posted in: evolution, prevention

