January 22, 2010
Mutations and the escape from immunity
If measles is caused by a virus and it can only be caught once in a life time, why do we catch the flu every year?
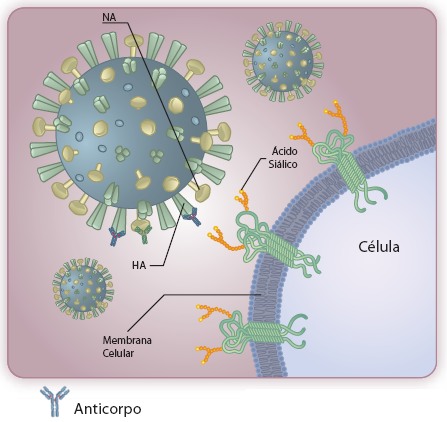
The antibody connected to Hemagglutinin (HA) prevents it from linking itself to the sialic acid of the cell.
When we have the flu, in a few days the body seems to get rid of the virus. The symptoms rarely last for two weeks and, in a higher period; it normally indicates complications caused by other microorganisms. This short period of the flu is due to our immune response.
As the virus replicates in the body of infected people, the immune system captures several pieces of the viral proteins, the so-called antigens, and produces antibodies against them. There are also other types of immune response but these do not cause long-term immunity, so they are not being considered. The two largest viral targets of the antibodies are the Hemagglutinin and Neuraminidase, since they are the most exposed proteins of the virus. When the antibodies bind to them, in addition to signaling to macrophages and other types of defense cells to attack the foreign body (the virus), it can still prevent the functioning of the virus. An antibody that attacks the region of recognition of the receptor of Hemagglutinin prevents it from binding to the cells. They are called neutralizing antibodies.
Thanks to this immune response, after a few days the Influenza is unable to infect us. But how is the virus able to return?
As previously seen, when it replicates its genome, the polymerase of the Influenza causes mutations. These mutations change the composition of the viral proteins. When the aminoacids (components of the proteins) from the region where the antibody bonds (antigen) are altered in consequence of the mutation, it may lose its affinity. Therefore, as the virus infects new hosts, it accumulates small changes that in the end result in the change of its antigens. This process is called gradual antigenic drift.
Mostly, these changes do not have to be sudden to have an effect. Changes in important locations for the recognition by the antibody of only one amino acid can be enough. This was proposed by one of the most important works carried out on the immunology of influenza that changed the way in which we study the virus.
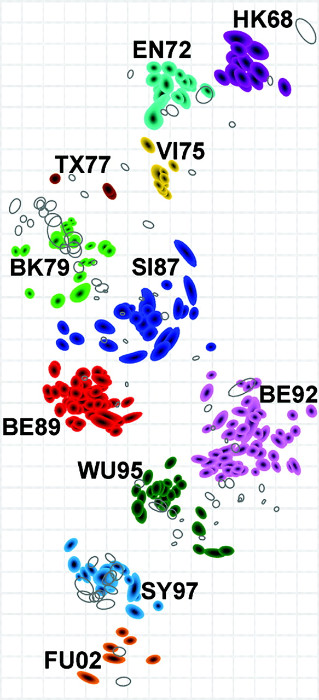
Antigenic map of hemagglutinin of H3N2 collected from 1968 (the year it infected humans) to 2003. The color of the circles defines the group to which that sample is related to. Each unit of distance between them represents the reduction of the immune response to the virus in relation to the antibodies produced against the previous one. Therefore, the greater the distance of one virus A from B, the smaller is its recognition by the antibodies against B.
Using an idea proposed in 2001, instead of only looking at the genetic difference between the samples of H3N2 causer of the human flu of different years, the authors decided to consider the immune response against them. Thus, maps are elaborated to point out how well the immune system recognizes that particular virus. The protein used in the study was the Hemagglutinin (HA) because it is against it that the most efficient antibodies are produced, reason why it is used in the annual vaccine.
Instead of a continuous change, the map showed that the virus tends to agglutinate in groups. Changes in sequences, sometimes, extensive may not represent a difference in the recognition by antibodies. On the other hand, in some cases, small changes in the HA are enough for a large antigenic distance. That is, with little mutations some Hemagglutinins can be much less recognized by the antibodies.
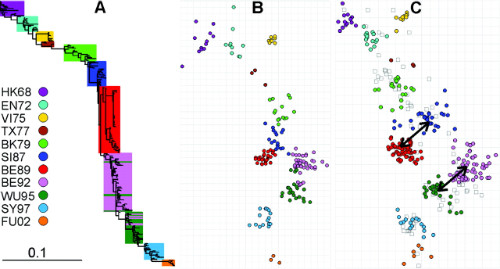
Genetic distance, in A and B, and antigenic, in C, isolated from H3N2. Some lineages, although genetically close, such as SI87 and BE89 in A and B, which only have one different amino acid, may not be antigenically close, as shown in C. This indicates that the antibodies produced against H3N2 in 1987 were not efficient against the 1989 virus.
It is also possible to see the change of the immune response in relation to the annual cycle. The groups remain dominant for three years in average, and the virus in this group normally appears around 2 years in advance and circulates for 2 more years after the dominance of the group.
Based on these results, the annual vaccines are seen differently. Today, the observation of the immune response to the virus of the seasonal vaccine and to the circulating virus, in addition to the genetic difference between them, is a very important factor in the production process. The research groups that monitor the Influenza and determine the lineages that will be part of the vaccine have to be carefully attentive to this fact because the change of an antigenic group of the Hemagglutinin of a virus, after it is being cultivated in eggs, can mean a failure in the immunization of a population.
Source:
Smith, D. (2004). Mapping the Antigenic and Genetic Evolution of Influenza Virus Science, 305 (5682), 371-376 DOI: 10.1126/science.1097211
7 Comments » Posted in: Immune response, fighting influenza, vaccine


[...] fact because the change of an antigenic group of the Hemagglutinin of a virus , … More: Mutations and the escape from immunity « Influenza A (H1N1) Blog Posted in H1N1 Flu, H1N1 Influenza Virus. Tags: and-determine, antigenic-group, because-the, [...]
[...] Antigenic map of hemagglutinin of H3N2 collected from 1968 (the year it … Continued here: Mutations and the escape from immunity « Influenza A (H1N1) Blog Posted in H3N2 Virus. Tags: carried-out, changed-the, collected-from, immunology, [...]
[...] is the original post: Mutations and the escape from immunity « Influenza A (H1N1) Blog Posted in H3N2 Flu Information. Tags: botucatu, caribbean, health, immune-response, [...]
[...] caused by other microorganisms. This short period of the flu … See original here: Mutations and the escape from immunity « Influenza A (H1N1) Blog Posted in H1N1 Virus Symptoms. Tags: body, normally-indicates, other-microorganisms-, period, [...]
[...] it normally indicates complications caused by other microorganisms. View original post here: Mutations and the escape from immunity « Influenza A (H1N1) Blog Posted in Flu Symptoms, H1N1 Flu Symptom Videos, H1N1 Symptoms. Tags: a-higher-period, for-two, [...]
this can’t be the whole story since unchange strains are also able
to infect again, although less likely.
And flu-B mutates at only half the rate than flu-A and still causes
yearly epidemics.
flu comes in waves, reaches a peak and then quickly declines
(although only ~10% are infected). The wave form is typical,
whether there is pre-existing immunity or not.
Another wave may happen 2-3 months later (tropics) but usually
only one wave per year in USA,Europe,Australia.
So the main driver must be some shortterm immunity.
And how can the wave regularly go down after ~5 weeks
when only 10% are infected ?
[...] We get measles once in our lifetimes but influenza much more frequently. Both viruses encode error-prone RNA polymerases, but the influenza glycoproteins are structurally more plastic than the measles counterparts – leading to escape from neutralizing antibodies. [...]